Mauve
<protect>
| Link/URL: | |
|---|---|
| What: |
Constructs multiple genome alignments in the presence of large-scale evolutionary events |
| Who: |
The University of Wisconsin - Madison |
| edit table |
</protect>
Contents
Short Description
Share your knowledge and ideas. How you can help. See Help pages if you need help with using the wiki
Mauve is a multiple-genome alignment program and visualizer.
"Mauve is a software
package that attempts to align orthologous and xenologous regions among two or more genome sequences that have undergone
both local and large-scale changes."[1]
Links
- Mauve at the Genome Evolution Laboratory: http://gel.ahabs.wisc.edu/mauve/
- Mauve-based tools
Requirements
Mauve pre-built binaries are available for Windows, Mac OS X (10.4+), and Linux. You can also download the source code and compile a version yourself.
User notes
Algorithm Summary
- Find local alignments (using multiMUMs)
- seed-and-extend
- can identify multi-MUMs in all genomes or in a subset of the genomes
- use the multiMUMs to generate a guide tree (phylogenetic tree)
- uses neighbor-joining
- use a subset of the multiMUMs to use as anchors (called LCBs)
- perform recursive anchoring to identifiy more alignment anchors
- preform a progressive alignment of each LCB using the guide tree
Definitions
In order to correctly express our ideas, we must use the correct terminology.
- MEM
- maximal exact match
- This is a local alignment that matches "exactly that cannot be extended 5' or 3' without encountering a mismatch." They are of length k.
- a multi-MEM is similar idea to a MEM, but for a multiple-sequence alignment instead of pairwise.
- MUM
- maximal unique match
- A MEM of at least length k bases that exists singly (uniquely) per genome.
- multi-MUMS are exactly matching subsequences shared by two or more genomes that occur only once in those genomes and that are bounded on either side by mismatches.
- Mauve uses a recursive anchoring strategy that iteratively reduces k to find multi-MUMs in regions that have undergone significant rearrangement/flux.
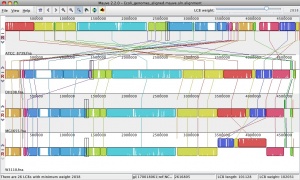
- LCB
- Locally colinear blocks
- Because recombination can cause genome rearrangements, orthologous regions of one genome may be reordered or inverted relative to another genome. During the alignment process, Mauve identifies conserved segments that appear to be internally free from genome rearrangements. Such regions are referred to as Locally Collinear Blocks (LCBs).
- The 'weight of the LCB is defined as the sum of the matches in that LCB. Mauve removes low-weight LCBs from a set of alignments before completion. (The default value of this cutoff is 3 times the minimum match size.) This weight provides a measure of confidence that the LCB is a true genome rearrangement rather than a spurious match.
- by specifying a high minimum LCB weight the user can "identify rearrangements that are very likely to exist"
- by specifying a low minimum LCB weight the user can trade some specificity for sensitivity to smaller genome rearrangements.
- LCBs described in the 2004 paper[2] are limited to sequence elements conserved among all the genomes being aligned.
- orphan
- In this context the word "orphan" means that a gene has no assigned orthology. This does not
orthologs
Mauve can export orthologs for annotated sequences. (Tools -> Export -> Export Orthologs)
The features that it can export are:
- gene
- CDS
- rRNA
- tRNA
- miscRNA
Programs
progressiveMauve
options
progressiveMauve usage: When each genome resides in a separate file: ./progressiveMauve [options] <seq1 filename> ... <seqN filename> When all genomes are in a single file: ./progressiveMauve [options] <seq filename> Options: --island-gap-size=<number> Alignment gaps above this size in nucleotides are considered to be islands [20] --profile=<file> (Not yet implemented) Read an existing sequence alignment in XMFA format and align it to other sequences or alignments --apply-backbone=<file> Read an existing sequence alignment in XMFA format and apply backbone statistics to it --disable-backbone Disable backbone detection --mums Find MUMs only, do not attempt to determine locally collinear blocks (LCBs) --seed-weight=<number> Use the specified seed weight for calculating initial anchors --output=<file> Output file name. Prints to screen by default --backbone-output=<file> Backbone output file name (optional). --match-input=<file> Use specified match file instead of searching for matches --input-id-matrix=<file> An identity matrix describing similarity among all pairs of input sequences/alignments --max-gapped-aligner-length=<number> Maximum number of base pairs to attempt aligning with the gapped aligner --input-guide-tree=<file> A phylogenetic guide tree in NEWICK format that describes the order in which sequences will be aligned --output-guide-tree=<file> Write out the guide tree used for alignment to a file --version Display software version information --debug Run in debug mode (perform internal consistency checks--very slow) --scratch-path-1=<path> Designate a path that can be used for temporary data storage. Two or more paths should be specified. --scratch-path-2=<path> Designate a path that can be used for temporary data storage. Two or more paths should be specified. --collinear Assume that input sequences are collinear--they have no rearrangements --scoring-scheme=<ancestral|sp_ancestral|sp> Selects the anchoring score function. Default is extant sum-of-pairs (sp). --no-weight-scaling Don't scale LCB weights by conservation distance and breakpoint distance --max-breakpoint-distance-scale=<number [0,1]> Set the maximum weight scaling by breakpoint distance. Defaults to 0.5 --conservation-distance-scale=<number [0,1]> Scale conservation distances by this amount. Defaults to 0.5 --muscle-args=<arguments in quotes> Additional command-line options for MUSCLE. Any quotes should be escaped with a backslash --skip-refinement Do not perform iterative refinement --skip-gapped-alignment Do not perform gapped alignment --bp-dist-estimate-min-score=<number> Minimum LCB score for estimating pairwise breakpoint distance --mem-clean Set this to true when debugging memory allocations --gap-open=<number> Gap open penalty --repeat-penalty=<negative|zero> Sets whether the repeat scores go negative or go to zero for highly repetitive sequences. Default is negative. --gap-extend=<number> Gap extend penalty --substitution-matrix=<file> Nucleotide substitution matrix in NCBI format --weight=<number> Minimum pairwise LCB score --min-scaled-penalty=<number> Minimum breakpoint penalty after scaling the penalty by expected divergence --hmm-p-go-homologous=<number> Probability of transitioning from the unrelated to the homologous state [0.00001] --hmm-p-go-unrelated=<number> Probability of transitioning from the homologous to the unrelated state [0.000000001] --hmm-identity=<number> Expected level of sequence identity among pairs of sequences, ranging between 0 and 1 [0.7] --seed-family Use a family of spaced seeds to improve sensitivity --disable-cache Disable recursive anchor search cacheing to workaround a crash bug --no-recursion Disable recursive anchor search Examples: ./progressiveMauve --output=my_seqs.xmfa my_genome1.gbk my_genome2.gbk my_genome3.fasta If genomes are in a single file and have no rearrangement: ./progressiveMauve --collinear --output=my_seqs.xmfa my_genomes.fasta
mauveAligner
The Mauve software shows the following message: (abbreviated)
MauveAligner is obsolete. it has been superseded by progressiveMauve.
options
Usage: ./mauveAligner [options] <seq1 filename> <sml1 filename> ... <seqN filename> <smlN filename> Options: --output=<file> Output file name. Prints to screen by default --mums Find MUMs only, do not attempt to determine locally collinear blocks (LCBs) --no-recursion Don't perform recursive anchor identification (implies --no-gapped-alignment) --no-lcb-extension If determining LCBs, don't attempt to extend the LCBs --seed-size=<number> Initial seed match size, default is log_2( average seq. length ) --max-extension-iterations=<number> Limit LCB extensions to this number of attempts, default is 4 --eliminate-inclusions Eliminate linked inclusions in subset matches. --weight=<number> Minimum LCB weight in base pairs per sequence --match-input=<file> Use specified match file instead of searching for matches --lcb-match-input Indicates that the match input file contains matches that have been clustered into LCBs --lcb-input=<file> Use specified lcb file instead of constructing LCBs (skips LCB generation) --scratch-path=<path> For large genomes, use a directory for storage of temporary data. Should be given two or more times to with different paths. --id-matrix=<file> Generate LCB stats and write them to the specified file --island-size=<number> Find islands larger than the given number --island-output=<file> Output islands the given file (requires --island-size) --backbone-size=<number> Find stretches of backbone longer than the given number of b.p. --max-backbone-gap=<number> Allow backbone to be interrupted by gaps up to this length in b.p. --backbone-output=<file> Output islands the given file (requires --island-size) --coverage-output=<file> Output a coverage list to the specified file (- for stdout) --repeats Generates a repeat map. Only one sequence can be specified --output-guide-tree=<file> Write out a guide tree to the designated file --collinear Assume that input sequences are collinear--they have no rearrangements Gapped alignment controls: --no-gapped-alignment Don't perform a gapped alignment --max-gapped-aligner-length=<number> Maximum number of base pairs to attempt aligning with the gapped aligner --min-recursive-gap-length=<number> Minimum size of gaps that Mauve will perform recursive MUM anchoring on (Default is 200) Signed permutation matrix options: --permutation-matrix-output=<file> Write out the LCBs as a signed permutation matrix to the given file --permutation-matrix-min-weight=<number> A permutation matrix will be written for every set of LCBs with weight between this value and the value of --weight Alignment output options: --alignment-output-dir=<directory> Outputs a set of alignment files (one per LCB) to a given directory --alignment-output-format=<directory> Selects the output format for --alignment-output-dir --output-alignment=<file> Write out an XMFA format alignment to the designated file Supported alignment output formats are: phylip, clustal, msf, nexus, mega, codon
- inGeno looks like a similar program. Not sure about the differences in methods, though.
--Daniel 19:11, 8 April 2009 (UTC)
See Also
References
See Help:References for how to manage references in EcoliWiki.
- ↑ Darling, AC et al. (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14 1394-403 PubMed
- ↑ http://genome.cshlp.org/content/14/7/1394.full