Category:Insertion Sequences
Contents
Insertion Sequences
list of sequences of all known IS elements in E. coli
General Information
Insertion sequences (IS) are distinct genetic units of less than 2.5kb of DNA whose only genes are related to transposition and self-propogation. [1] [2] IS elements are capable of independently replicating and inserting into new sites on the genome without leaving the original site and have short (10-40bp) inverted repeats at both termini. [3]
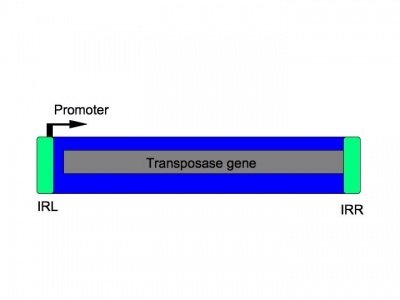
Figure 1. Typical organization of IS Elements from Mobile DNA II (pp 307).[2] IRR is the right inverted repeat & IRL is the left inverted repeat.
E. coli Insertion Sequences
Mobile insertion elements are involved in insertions, deletions and inversions of the chromosome. In E. coli, most are 700-2000 bp in length and vary in their transpositional activity. Homologous recombination (mediated by RecA) can occur between IS elements, and this contributes to the plasticity of the bacterial genome.
Cointegration and Hfr strains of E. coli
Cointegration is the process by which two circular replicons fuse to create a single replicon. Cointegration can occur at preexisting copies of the same IS element on the chromosome and F plasmid, resulting in the creation of an Hfr strain. [4]
Naming conventions for IS Elements in E. coli
IS elements typically are denoted by a numerical index, such as IS4. Each IS element contains a set of genes, designated by a letter, such as insA. IS elements typically encode 1-3 proteins[5].
E. coli proteins involved in transposition
The proteins involved in transposition vary depending on the IS element. There are binding sites for IHF in IS1 [6] , IS903 and IS10[7] [2].
See also
- Cis Elements
- Transposons
- Mobile Genetic Elements
- list of sequences of all known IS elements in E. coli
External links
- IS Finder - About IS Finder on EcoliWiki
- Transposase structures from PDB
- Transposase activity definition from GO
- Transposition process definition from GO
__SUBCATEGORY_SECTION__
__PAGE_SECTION__
References
See Help:References for how to manage references in EcoliWiki.
- ↑ Neidhardt, FC et al. (1996) Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology 2nd ed. (ASM Press, Washington, DC)
- ↑ 2.0 2.1 2.2 Edited by Craig, Nancy L., Robert Craigie, Martin Gellert and Alan M. Lambowitz (2002) Mobile DNA II (ASM Press, Washington, D.C.) ISBN 1-55581-209-0
- ↑ Shapiro, James A., 'Mobile Genetic Elements.' Academic Press Inc. 1983-4. ISBN 0-12-638680-3
- ↑ Ohtsubo, E et al. (1981) Mechanism of insertion and cointegration mediated by IS1 and Tn3. Cold Spring Harb. Symp. Quant. Biol. 45 Pt 1 283-95 PubMed
- ↑ Shapiro, James A., 'Mobile Genetic Elements.' Academic Press Inc. 1983-4. ISBN 0-12-638680-3
- ↑ Gamas, P et al. () DNA sequence at the end of IS1 required for transposition. Nature 317 458-60 PubMed
- ↑ Huisman, O et al. (1989) Mutational analysis of IS10's outside end. EMBO J. 8 2101-9 PubMed
Pages in category "Insertion Sequences"
The following 53 pages are in this category, out of 53 total.
I
- IS1
- IS1203
- IS1294
- IS1397
- IS1414
- IS150
- IS186
- IS2
- IS200
- IS26
- IS3
- IS30
- IS3000
- IS3411
- IS4
- IS5
- IS50
- IS5075
- IS600
- IS609
- IS621
- IS679
- IS682
- IS903
- IS91
- IS911
- ISEc1
- ISEc10
- ISEc11
- ISEc12
- ISEc13
- ISEc14
- ISEc15
- ISEc16
- ISEc17
- ISEc18
- ISEc19
- ISEc20
- ISEc21
- ISEc22
- ISEc23
- ISEc24
- ISEc25
- ISEc26
- ISEc27
- ISEc30
- ISEc31
- ISEc32
- ISEc33
- ISEc8
- ISEc9
- ISEcB1
- ISEcp1