ompA:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
ompA |
|---|---|
| Gene Synonym(s) |
ECK0948, b0957, JW0940, con, tolG, tut[1], tut |
| Product Desc. |
outer membrane protein 3a (II*;G;d)[2][3] Outer membrane protein A; Outer membrane porin A; T-even phage receptor; weak porin; homodimer, abundant cell surface protein[4] Discovered to influence biofilm formation[5] OmpA inhibits cellulose production through the CpxRA stress response system, and this reduction in cellulose increases biofilm formation on hydrophobic surfaces[6]. Deletion of ompA increases cellulose which decreases biofilm formation on hydrophobic surfaces[6]. |
| Product Synonyms(s) |
outer membrane protein A (3a;II*;G;d)[1], B0957[2][1], Tut[2][1], Con[2][1], TolG[2][1], OmpA[2][1], con, ECK0948, JW0940, tolG, tut, b0957 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
OmpA is required for colicins K and L and the stabilization of mating aggregates. OmpA is elevated 5X during growth at 4C (enabled by Oleispira antarctica Cpn10/60 transgene). OmpA expression is elevated during biofilm formation (Orme, 2006). OmpA is required for biofilm formation under some condtions. Translation of OmpF is inhibited by the rseX sRNA. Alkali-inducible Binds TrxA (Kumar, 2004). Perhaps because it is so abundant, OmpA is found in some inner membrane preparations.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
ompA |
|---|---|
| Mnemonic |
Outer membrane protein |
| Synonyms |
ECK0948, b0957, JW0940, con, tolG, tut[1], tut |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
21.95 minutes |
MG1655: 1019276..1018236 |
||
|
NC_012967: 1037144..1036104 |
||||
|
NC_012759: 921204..922244 |
||||
|
W3110 |
|
W3110: 1020475..1019435 |
||
|
DH10B: 1073204..1072164 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
1018239 |
Edman degradation |
PMID:340230 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ompA(del) (Keio:JW0940) |
deletion |
deletion |
PMID:16738554 |
||||
|
ompA::Tn5KAN-2 (FB20072) |
Insertion at nt 518 in Minus orientation |
PMID:15262929 |
does not contain pKD46 | ||||
|
ompA::kan |
deletion |
Biolog:respiration |
unable to respire Acetate |
PMID:16095938 |
|||
|
ompA252 |
|||||||
|
ompA256 |
|||||||
|
ompA772(del)::kan |
PMID:16738554 |
||||||
|
P678-54ompA2101 |
Sensitivity to |
increased sensitivity to acid, table 3. |
PMID:1314797 |
Experimental strain: RJR200 |
|||
|
ompA in strain P460 |
Resistant to |
Resistant to bacteriocin Colicin K |
PMID:1991710 |
Strain: P460 |
table 3 | ||
|
ompA in strain P1636 |
Resistant to |
Resistant to bacteriocin Colicin L |
PMID:1991710 |
Strain: P460 |
table 3 | ||
|
ompA in strain P460 |
Resistant to |
Resistant to bacteriophage K3 |
PMID:1991710 PMID:6819426 |
Strain: p460 |
Figure 6 | ||
|
ompA1 |
Resistant to |
Resistant to novobicin |
PMID:6819426 |
Strains: ompA1, ompA2 and ompA3 |
table 6 | ||
|
ompA1 |
Resistant to |
Resistant to chelator, specifically EDTA |
PMID:6819426 |
Strains: ompA1, ompA2 and ompA3 |
table 6 | ||
|
ompA in strain CC205 |
Resistant to |
Resistant to Phage TUII* |
PMID:6819426 |
Strain: CC205 |
Table 4 | ||
|
ompA in strain CC205 |
Resistant to |
Resistant to phage K3hl |
PMID:6819426 |
strain CC205 |
Table 4 | ||
|
ompA in strain CC205 |
Resistant to |
Resistant to pphage K3 |
PMID:6819426 |
Strain: cc205 |
table 4 | ||
|
ompA in strain P460 |
Resistant to |
Resistance to Bacteriocin JF246 |
PMID:783128 |
Strain P460 |
Table 3 | ||
|
con-tsx in P1731 |
Resistant to |
Resistance to Bacteriophage T6 |
PMID:786287 |
Double Mutation to the con & tsx genes cause resistance to the phage T6, possibly due to the reduced production of a protein (tsx-protein) thought to be involved phage binding to the cell. | |||
|
con-tsx in P1731 |
Resistant to |
Resistance to Colicin K |
PMID:786287 |
Double Mutation to the con & tsx genes cause resistance to the Colicin K, possibly due to the reduced production of a protein (tsx-protein) thought to be involved phage binding to the cell. | |||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW0940 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCAAAAAGACAGCTATCGCGAT Primer 2:CCAGCCTGCGGCTGAGTTACAA | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 57% [8] | ||
|
Linked marker |
est. P1 cotransduction: 14% [8] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10669 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10669 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000662 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0663 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0003240 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
OmpA |
|---|---|
| Synonyms |
outer membrane protein A (3a;II*;G;d)[1], B0957[2][1], Tut[2][1], Con[2][1], TolG[2][1], OmpA[2][1], con, ECK0948, JW0940, tolG, tut, b0957 |
| Product description |
outer membrane protein 3a (II*;G;d)[2][3] Outer membrane protein A; Outer membrane porin A; T-even phage receptor; weak porin; homodimer, abundant cell surface protein[4] Discovered to influence biofilm formation[5] OmpA inhibits cellulose production through the CpxRA stress response system, and this reduction in cellulose increases biofilm formation on hydrophobic surfaces[6]. Deletion of ompA increases cellulose which decreases biofilm formation on hydrophobic surfaces[6]. |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0000746 |
conjugation |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0184 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005198 |
structural molecule activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002368 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005515 |
protein binding |
PMID:9701827 |
IDA: Inferred from Direct Assay |
F |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0005515 |
protein binding |
PMID:12509434 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0005886 |
plasma membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-1003 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
PMID:12509434 |
IDA: Inferred from Direct Assay |
C |
complete | |||
|
GO:0006810 |
transport |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0813 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009279 |
cell outer membrane |
PMID:12509434 |
IDA: Inferred from Direct Assay |
C |
complete | |||
|
GO:0006811 |
ion transport |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0406 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0019867 |
outer membrane |
PMID:12509434 |
IDA: Inferred from Direct Assay |
C |
complete | |||
|
GO:0009279 |
cell outer membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000498 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0015288 |
porin activity |
PMID:10636850 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0009279 |
cell outer membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002368 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
PMID:3047121 |
IDA: Inferred from Direct Assay |
C |
complete | |||
|
GO:0009279 |
cell outer membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006664 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006810 |
transport |
PMID:10636850 |
IDA: Inferred from Direct Assay |
P |
complete | |||
|
GO:0009279 |
cell outer membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006665 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009279 |
cell outer membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006690 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046718 |
entry of virus into host cell |
PMID:367782 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0000746 |
conjugation |
PMID:3311813 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0009279 |
cell outer membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011250 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009597 |
detection of virus |
PMID:367782 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0006811 |
ion transport |
PMID:10636850 |
IDA: Inferred from Direct Assay |
P |
complete | |||
|
GO:0019867 |
outer membrane |
PMID:3047120 |
IDA: Inferred from Direct Assay |
C |
complete | |||
|
GO:0009279 |
cell outer membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0998 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009279 |
cell outer membrane |
PMID:3047120 |
IDA: Inferred from Direct Assay |
C |
complete | |||
|
GO:0009597 |
detection of virus |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0580 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
PMID:12509434 |
IDA: Inferred from Direct Assay |
C |
complete | |||
|
GO:0016021 |
integral to membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000498 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002368 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005515 |
protein binding |
PMID:9701827 |
IDA: Inferred from Direct Assay |
F |
with TolB |
complete | ||
|
GO:0016021 |
integral to membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006664 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009279 |
cell outer membrane |
PMID:20081027 |
IDA: Inferred from Direct Assay |
C |
complete | |||
|
GO:0016021 |
integral to membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006690 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011250 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0812 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-9909 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046718 |
entry of virus into host cell |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0580 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046930 |
pore complex |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0626 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006974 |
response to DNA damage stimulus |
PMID:11967071 |
IEP: Inferred from Expression Pattern |
P |
complete | |||
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
nuoC |
PMID:19402753 |
MALDI(Z-score):25.885148 | |
|
Protein |
rplI |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
clpB |
PMID:19402753 |
LCMS(ID Probability):99.3 | |
|
Protein |
pflB |
PMID:19402753 |
MALDI(Z-score):21.041644 | |
|
Protein |
tig |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rplW |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rpsN |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
Hfq |
Hfq stimulates ompA mRNA degradation in a growth rate-dependent manner |
PMID:11804582 PMID 9826663 |
Hfq interferes with ribosome binding, exposing mRNA to endonuclease cleavage |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes | |
|---|---|---|---|---|---|
|
outer membrane |
From EcoCyc[3] |
||||
|
Outer Membrane |
PMID:340230, PMID:7001461, PMID:8740179, PMID:9298646, PMID:9629924. |
||||
|
Outer Membrane |
PMID:9298646 |
| |||
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MKKTAIAIAV ALAGFATVAQ AAPKDNTWYT GAKLGWSQYH DTGFINNNGP THENQLGAGA FGGYQVNPYV GFEMGYDWLG RMPYKGSVEN GAYKAQGVQL TAKLGYPITD DLDIYTRLGG MVWRADTKSN VYGKNHDTGV SPVFAGGVEY AITPEIATRL EYQWTNNIGD AHTIGTRPDN GMLSLGVSYR FGQGEAAPVV APAPAPAPEV QTKHFTLKSD VLFNFNKATL KPEGQAALDQ LYSQLSNLDP KDGSVVVLGY TDRIGSDAYN QGLSERRAQS VVDYLISKGI PADKISARGM GESNPVTGNT CDNVKQRAAL IDCLAPDRRV EIEVKGIKDV VTQPQA |
| Length |
346 |
| Mol. Wt |
37.2 kDa |
| pI |
6.4 (calculated) |
| Extinction coefficient |
52,830 - 53,080 (calc based on 17 Y, 5 W, and 2 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0003240 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10669 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10669 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000662 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0663 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
2.99E+04 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
E. coli K-12 EMG2 |
140 |
molecules/cell |
|
Quantitative protein sequencing |
Spot ID: Mll8-2 |
PMID: 9298646 |
|
Protein |
E. coli K-12 EMG2 |
2300 |
molecules/cell |
|
Quantitative protein sequencing |
Spot ID: MB |
PMID: 9298646 |
|
Protein |
E. coli K-12 EMG2 |
3380 |
molecules/cell |
|
Quantitative protein sequencing |
Spot ID: M52�2 |
PMID: 9298646 |
|
Protein |
E. coli K-12 EMG2 |
1480 |
molecules/cell |
|
Quantitative protein sequencing |
Spot ID: Ml51 |
PMID: 9298646 |
|
Protein |
E. coli K-12 MG1655 |
207618 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
69396 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
106768 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
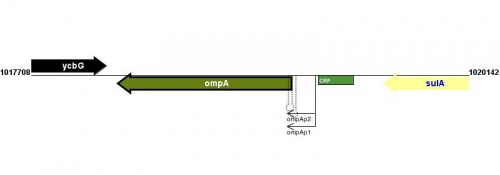 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:1019256..1019296
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for ompA | |
|
microarray |
Summary of data for ompA from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (1019201..1019676) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to ompA Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10669 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0663 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10669 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000662 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0003240 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
OMPA |
From SHIGELLACYC |
|
E. coli O157 |
OMPA |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10669 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10669 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000662 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0663 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0003240 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 Barrios, AF et al. (2006) Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol. Bioeng. 93 188-200 PubMed
- ↑ 6.0 6.1 6.2 6.3 Ma, Q & Wood, TK (2009) OmpA influences Escherichia coli biofilm formation by repressing cellulose production through the CpxRA two-component system. Environ. Microbiol. 11 2735-46 PubMed
- ↑ 7.0 7.1 7.2 CGSC: The Coli Genetics Stock Center
- ↑ 8.0 8.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories


